4. Membrane Processes Layouts#
In this set of notes we will use an example ultrafiltration process to introduce material balances in different process configurations. We will consider a Batch, semi-Batch (feed and bleed) and continuous membrane processes to develop general ideas around material balances in membrane units. Every example will be introduced by a slight variant of a common problem statement.
4.1 Batch#
Problem Statement
500 l of fruit juice are concentrated from an initial solid-residue content of 0.05 kg/l to a solid-residue content of 0.2 kg/l through a batch microfiltration process. The total area of the membrane is of 20 \(\mathrm{m^2}\) and the flux of pure water through the membrane is captured by the empirical expression: \(J=BC^{-1} [m/h]\) with \(B=0.1\) where C is the solid residue concentration in kg/l. Compute the process time necessary to reach the target concentration.
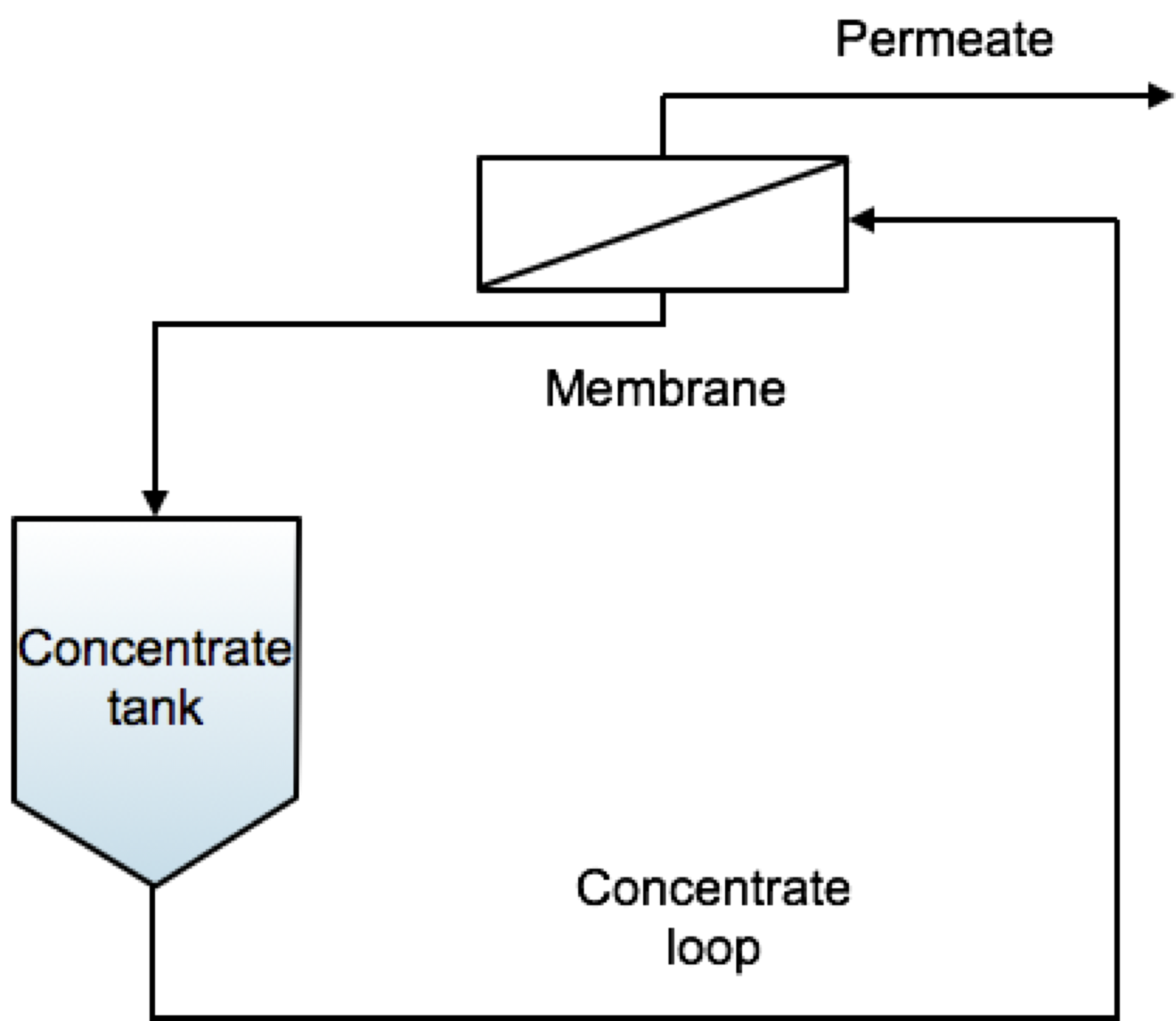
Fig. 1 Schematics of a Batch membrane process.#
Solution trace
The global differential material balance (volume basis) reads:
where \(V\) is the volume in the concentrate loop, \(J\) the permeate flux per unit area and \(A\) the total membrane area. The permeate flux is defined by the expression:
with \(B=0.1\).
Since the solid residue never leaves the retentate loop, the differential material balance on the solid residue reads:
Which means that the total mass of solid residue is constant and equal to its initial mass:
The global differential material balance can thus be rewritten as:
In this simple case it can be integrated analytically to compute the volume as a function of time:
Putting together both sides of the equation and substituting \(V^\prime\) with \(V\) for the sake of simplicity in the notation we get:
which describes the time dependence of the volume in the batch membrane separator.
The concentration can thus be calculated as:
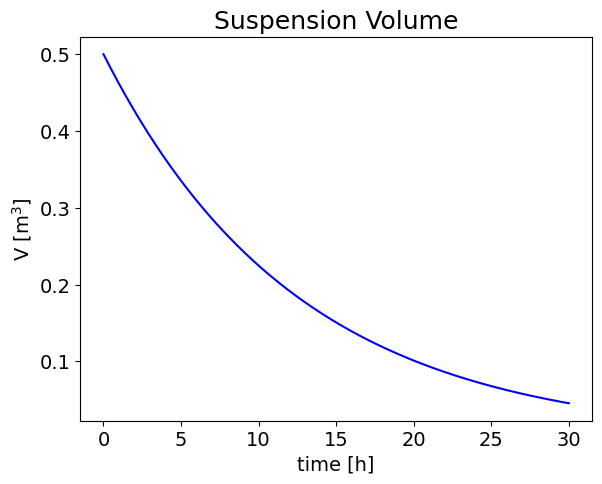
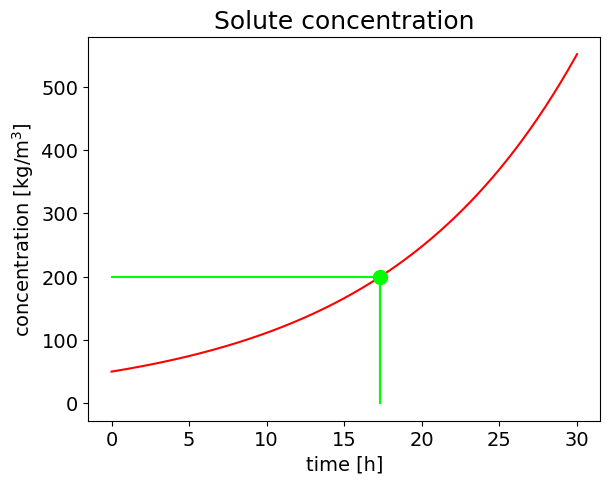
The process time necessary to obtain a solid residue concentration of 0.2 \([kg/l]\) is of 17.3 \([h]\)
Numerical solution
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import fsolve
# Parameters:
N = 500 #number of points
time = np.linspace(0, 30, N)
#Every length in m
A=20; #m^2
B=0.1;
C0=0.05*1E3; #kg /l * dm^3/m^3
V0=500*1E-3; #l * m^3/dm^3
C_specific = 0.2*1E3
#Operating Equation
C = C0*np.exp(B*A/V0/C0*time)
V = V0*np.exp(-B*A/V0/C0*time)
def equation(proc_time):
eq1 = C0*np.exp(B*A/V0/C0*proc_time) - C_specific
return eq1
process_time = fsolve(equation,1)
#Plotting
figure=plt.figure()
axes = figure.add_axes([0.1,0.1,0.8,0.8])
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
axes.plot(time,V, marker=' ' , color='b')
plt.title('Suspension Volume', fontsize=18);
axes.set_xlabel('time [h]', fontsize=14);
axes.set_ylabel('V [m$^3$]',fontsize=14);
figure=plt.figure()
axes = figure.add_axes([0.1,0.1,0.8,0.8])
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
axes.plot(time,C, marker=' ' , color='r')
axes.plot(np.array([process_time[0],process_time[0]]),np.array([0, C_specific]), marker=' ' , color='lime', markersize=3)
axes.plot(np.array([0,process_time[0]]),np.array([C_specific, C_specific]), marker=' ' , color='lime', markersize=3)
axes.plot([process_time,process_time],[0, C_specific], marker=' ' , color='lime', markersize=3)
axes.plot(process_time,C_specific, marker='o' , color='lime', markersize=10)
plt.title('Solute concentration', fontsize=18);
axes.set_xlabel('time [h]', fontsize=14);
axes.set_ylabel('concentration [kg/m$^3$]',fontsize=14);
print("The process time necessary to obtain a solid residue concentration of", C_specific, "[kg/l] is:", process_time, "[h]")
The process time necessary to obtain a solid residue concentration of 200.0 [kg/l] is: [17.32867951] [h]
4.2 Feed and Bleed#
Feed and Bleed: Problem Statement
500 l of fruit juice are concentrated in 5 h of steady state operation from an initial solid-residue content of 0.05 \(kg/l\) through a feed and bleed process. The total area of the membrane is of 20 \(m^2\) and the flux of pure water through the membrane is captured by the empirical expression: \(J=BC^{-1} [m/h]\) with \(B=0.1\) where C is the solid residue concentration in kg/l. Is the steady state concentration of the retentate compatible with the specifics of 0.2 \(kg/l\)?
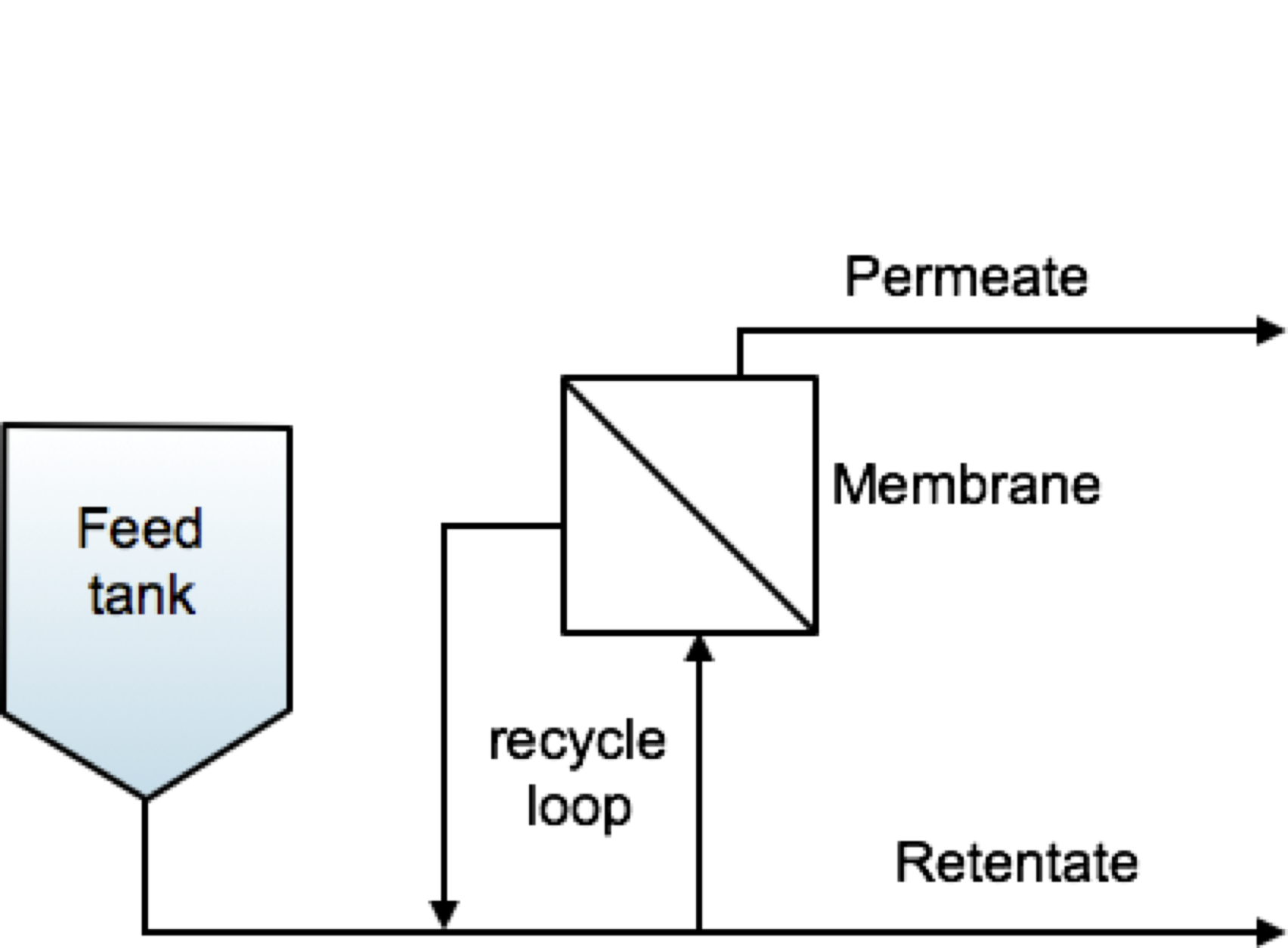
Fig. 2 Feed and Bleed process configuration scheme#
Feed and Bleed: Solution trace
Also in this case the global differential material balance and the solid-residue material balance can be written as follows:
At steady state \({dV}/{dt}=0\) as well as \({dm}/{dt}=0\). The two ODEs become then two algebraic equations that should be solved together to compute the steady state concentration. From the global material balance we get:
and then, with some manipulations:
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import fsolve
# data:
A=20; #[m^2]
B=0.1;
C0=0.05*1E3; #kg /l * dm^3/m^3
V0=500*1E-3; #l * m^3/dm^3
process_time=5; # [h]
Cout=C0+A*B*process_time/V0
print("The steady state concentration is", Cout, "[kg/m^3]")
The steady state concentration is 70.0 [kg/m^3]
4.3 Cascade configuration#
Problem Statement
500 l of fruit juice are concentrated in 5 h of steady state operation from an initial solid-residue content of 0.05 kg/l through four membrane separation units characterised by a total membrane area of 20 \(\mathrm{m^2}\) each. The flux of pure water through the membrane is captured by the empirical expression: \(J=BC^{-1} [m/h]\) with \(B=0.1\) where C is the solid residue concentration in kg/l.
Is it more efficient to design a single stage configuration with four units in parallel or a cascade configuration with four units in series?
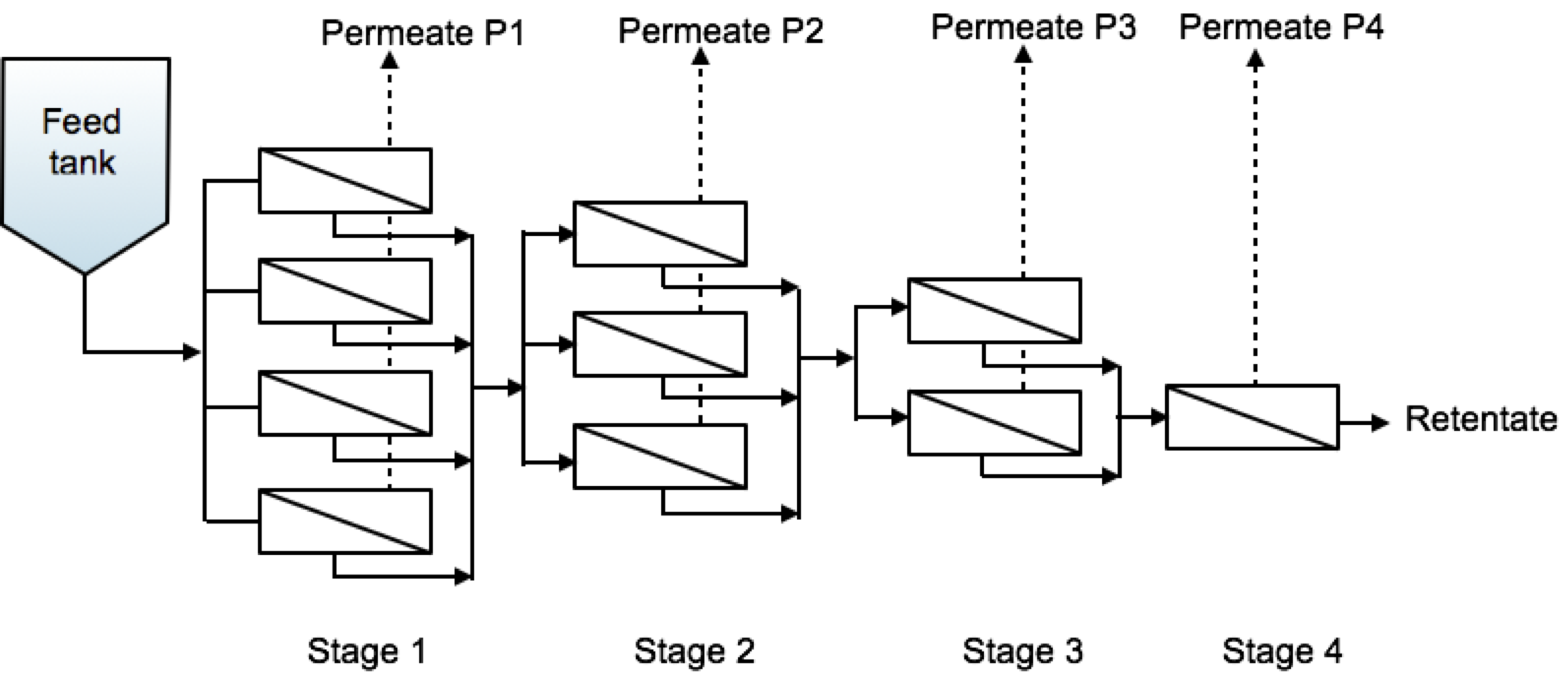
Fig. 3 Cascade configuration schematic representation.#
Cascade: Solution trace
Each stage can be treated like a single unit in which the feed stream corresponds to the retentate stream from the previous unit. We can thus define:
where the index \(i\) identifies the stage.
The material balances for each stage at steady state can thus be written as:
where \(n_i\) is the number of modules used in stage \(i\).
For each stage we can thus compute the steady state concentration and concentrate volumetric flow by solving sequentially the following equations:
with \(i=1...N\) with \(N\) is the total number of stages.
Numerical Solution
The solution of a cascade composed of any number of stages, each formed by an arbitrary number of modules in parallel can be tackled sequentially through a simple cycle similar to the following, let’s for example consider a system that reflects the sketch above i.e. with 4 stages implementing 4, 3, 2, and 1 modules each:
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import fsolve
# data:
A=20;
B=0.1;
C0=0.05*1E3; #kg /l * dm^3/m^3
V0=500*1E-3; #l * m^3/dm^3
process_time=5; # [h]
# The number of elements of this array corresponds to the number of stages.
# The value in each element is the number of modules per stage.
n=np.array([4,3,2,1]);
CIN=np.append(C0,np.zeros(np.size(n)-1))
FIN=np.append(V0/process_time, np.zeros(np.size(n)-1));
# Input to the intermediate stages
for i in range(1,np.size(n)):
CIN[i]=CIN[i-1]+n[i-1]*A*B/FIN[i-1];
FIN[i]=CIN[i-1]*FIN[i-1]/CIN[i];
# Output concentration
Cout=CIN[np.size(n)-1]+n[np.size(n)-1]*A*B/FIN[np.size(n)-1];
print("The steady state concentration is", Cout, "[kg/m^3]")
The steady state concentration is 720.7199999999999 [kg/m^3]
In order to answer the problem request one should solve the system for two different configurations. In the first, representing a single-stage configuration with four membrane units in parallel, the number of stages should be set to \(N=1\) and the number of units in the first stage \(n_1\) to 4.
# The number of elements of this array corresponds to the number of stages.
# The value in each element is the number of modules per stage.
n=np.array([4]);
CIN=np.append(C0,np.zeros(np.size(n)-1))
FIN=np.append(V0/process_time, np.zeros(np.size(n)-1));
# Input to the intermediate stages
for i in range(1,np.size(n)):
CIN[i]=CIN[i-1]+n[i-1]*A*B/FIN[i-1];
FIN[i]=CIN[i-1]*FIN[i-1]/CIN[i];
# Output concentration
Cout=CIN[np.size(n)-1]+n[np.size(n)-1]*A*B/FIN[np.size(n)-1];
print("The steady state concentration is", Cout, "[kg/m^3]")
The steady state concentration is 130.0 [kg/m^3]
In the second the number of stages should be set to \(N=4\), each of the stages being assembled as a single unit (\(n_i=1\) for \(i=[1, 4]\)).
# The number of elements of this array corresponds to the number of stages.
# The value in each element is the number of modules per stage.
n=np.array([1, 1, 1, 1]);
CIN=np.append(C0,np.zeros(np.size(n)-1))
FIN=np.append(V0/process_time, np.zeros(np.size(n)-1));
# Input to the intermediate stages
for i in range(1,np.size(n)):
CIN[i]=CIN[i-1]+n[i-1]*A*B/FIN[i-1];
FIN[i]=CIN[i-1]*FIN[i-1]/CIN[i];
# Output concentration
Cout=CIN[np.size(n)-1]+n[np.size(n)-1]*A*B/FIN[np.size(n)-1];
print("The steady state concentration is", Cout, "[kg/m^3]")
The steady state concentration is 192.07999999999998 [kg/m^3]
The cascade configuration allows for a more efficient process since it allows to obtain a larger concentration with the same number of modules.
